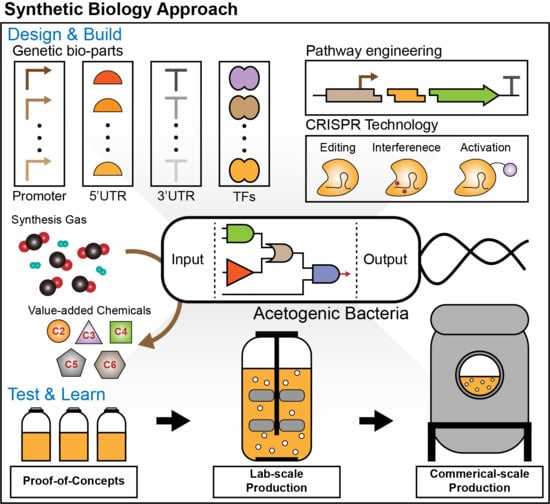
Synthetic Biology on Acetogenic Bacteria for Highly Efficient Conversion of C1 Gases to Biochemicals
Abstract
:1. Introduction
2. Development of Genetic Manipulation Tools in Acetogens
2.1. Development of Plasmid-Based Engineering Tools in Acetogens
2.2. Development of Genome Engineering Tools in Acetogens
2.2.1. ClosTron
2.2.2. Transposon Mutagenesis
2.2.3. Homologous Recombination
2.2.4. CRISPR-Cas Genome Editing
3. Engineering of Acetogens for Biochemical Production
3.1. Improvement of C1-Fixing Pathways in Acetogens
3.1.1. Engineering of the WL Pathway
3.1.2. Discovery of Novel C1 Gas Fixing Pathways
3.2. Production of Native Biochemicals
3.2.1. Acetate
3.2.2. Ethanol
3.2.3. 2,3-Butanediol (2,3-BDO)
3.3. Production of Non-Native Biochemicals
3.3.1. C3: Acetone or Isopropanol
3.3.2. C4: n-Butanol or Butyrate
3.3.3. C5: Isoprene
3.3.4. Poly-3-hydroxybutyrate (PHB)
4. Synthetic Biology Approach on Acetogens
4.1. Development of Synthetic Biology Tools in Acetogens
4.1.1. Application of Multi-Omics Data with Genome-Scale Metabolic Models
4.1.2. Development of Synthetic Promoters in Acetogens
4.2. Transcriptional Regulation System Based on CRISPR-Cas in Acetogens
4.2.1. Repurposing CRISPR-Cas System for Metabolic Engineering
4.2.2. Genome-Wide CRISPRi/a Screening for Functional Genomic Studies
4.2.3. CRISPRi-Based Synthetic Genetic Circuits
5. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| 2,3-BDO | 2,3-Butanediol |
| 5-FOA | 5-fluoroorotic acid |
| ACK | Acetate kinase |
| AOR | Aldehyde-ferredoxin oxidoreductase |
| ATP | Adenosine triphosphate |
| Att/Int | Attachment/Integration |
| Cas | CRISPR-associated protein |
| CO | Carbon monoxide |
| CO2 | Carbon dioxide |
| CODH/ACS | Carbon monoxide dehydrogenase/acetyl-CoA synthase |
| CRISPR | Clustered Regularly Interspaced Short Palindromic Repeats |
| CRISPRa | CRISPR activation |
| CRISPRi | CRISPR interference |
| crRNA | CRISPR RNA |
| tracrRNA | Trans-activating CRISPR RNA |
| dCas | Deactivated Cas9 |
| DNA | DeoxyriboNucleic Acid |
| DSB | Double strans break |
| DXS | 1-deoxyxylulose 5-phosphate synthase |
| FbFP | Flavin mononucleotide based fluorescent protein |
| FMN | Flavin mononucleotide |
| GEM | Genome-scale metabolic model |
| GSRP | Glycine synthase-reductase |
| HR | Homologous recombination |
| IPP | Isopentenyl pyrophosphate |
| ITRs | Inverted terminal repeats |
| LHA | Left homology arm |
| LOV | Light-oxgen-voltage sensing domain |
| MEK | Methyl-ethyl ketone |
| MVA | Mevlaonic acid |
| NAD/NADH | Nicotinamide adenine dinucleotide |
| NADP/NADPH | Nicotinamide adenine dinucleotide phosphate |
| NGS | Next-generation sequencing |
| PAM | Protospacer adjacent motif |
| PHA | Polyhydroxyalkanoate |
| PHB | Poly-3-hydroxybutyrate |
| RBS | Ribosome-binding sites3 |
| RHA | Right homology arm |
| RNA | RiboNucleic Acid |
| sgRNA | Single guide RNA |
| THF | Tetrahydrofolate |
| TSS | Transcription start site |
| UTRs | Untranslated regions |
| WL | Wood-Ljungdahli |
References
- Min, F.; Dennis, S.; Kopke, M. Gas Fermentation for Commercial Biofuels Production. In Liquid, Gaseous and Solid Biofuels—Conversion Techniques; IntechOpen: London, UK, 2013. [Google Scholar] [CrossRef] [Green Version]
- Schiel-Bengelsdorf, B.; Dürre, P. Pathway engineering and synthetic biology using acetogens. FEBS Lett. 2012, 586, 2191–2198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liew, F.; Martin, M.E.; Tappel, R.C.; Heijstra, B.D.; Mihalcea, C.; Köpke, M. Gas Fermentation—A Flexible Platform for Commercial Scale Production of Low-Carbon-Fuels and Chemicals from Waste and Renewable Feedstocks. Front. Microbiol. 2016, 7, 694. [Google Scholar] [CrossRef] [PubMed]
- Schuchmann, K.; Müller, V. Autotrophy at the thermodynamic limit of life: A model for energy conservation in acetogenic bacteria. Nat. Rev. Genet. 2014, 12, 809–821. [Google Scholar] [CrossRef]
- Bengelsdorf, F.R.; Straub, M.; Dürre, P. Bacterial synthesis gas (syngas) fermentation. Environ. Technol. 2013, 34, 1639–1651. [Google Scholar] [CrossRef] [PubMed]
- Köpke, M.; Mihalcea, C.; Bromley, J.C.; Simpson, S.D. Fermentative production of ethanol from carbon monoxide. Curr. Opin. Biotechnol. 2011, 22, 320–325. [Google Scholar] [CrossRef]
- Köpke, M.; Mihalcea, C.; Liew, F.; Tizard, J.H.; Ali, M.S.; Conolly, J.J.; Al-Sinawi, B.; Simpson, S.D. 2,3-Butanediol Production by Acetogenic Bacteria, an Alternative Route to Chemical Synthesis, Using Industrial Waste Gas. Appl. Environ. Microbiol. 2011, 77, 5467–5475. [Google Scholar] [CrossRef] [Green Version]
- Köpke, M.; Simpson, S.D. Pollution to products: Recycling of ’above ground’ carbon by gas fermentation. Curr. Opin. Biotechnol. 2020, 65, 180–189. [Google Scholar] [CrossRef]
- Latif, H.; Zeidan, A.A.; Nielsen, A.T.; Zengler, K. Trash to treasure: Production of biofuels and commodity chemicals via syngas fermenting microorganisms. Curr. Opin. Biotechnol. 2014, 27, 79–87. [Google Scholar] [CrossRef] [Green Version]
- Heijstra, B.D.; Leang, C.; Juminaga, A. Gas fermentation: Cellular engineering possibilities and scale up. Microb. Cell Fact. 2017, 16, 60. [Google Scholar] [CrossRef] [Green Version]
- Cameron, D.E.; Bashor, C.J.; Collins, J.J. A brief history of synthetic biology. Nat. Rev. Genet. 2014, 12, 381–390. [Google Scholar] [CrossRef]
- Zhang, W.; Nielsen, D.R. Synthetic biology applications in industrial microbiology. Front. Microbiol. 2014, 5, 451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, L.; Zhu, Y.; Zhang, Y.; Li, Y. Engineering the robustness of industrial microbes through synthetic biology. Trends Microbiol. 2012, 20, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Purnick, P.E.M.; Weiss, R. The second wave of synthetic biology: From modules to systems. Nat. Rev. Mol. Cell Biol. 2009, 10, 410–422. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, L.; Li, J.; Du, G.; Chen, J. Synthetic Biology Toolbox and Chassis Development in Bacillus subtilis. Trends Biotechnol. 2019, 37, 548–562. [Google Scholar] [CrossRef]
- Chi, H.; Wang, X.; Shao, Y.; Qin, Y.; Deng, Z.; Wang, L.; Chen, S. Engineering and modification of microbial chassis for systems and synthetic biology. Synth. Syst. Biotechnol. 2019, 4, 25–33. [Google Scholar] [CrossRef]
- Choi, K.R.; Jang, W.D.; Yang, D.; Cho, J.S.; Park, D.; Lee, S.Y. Systems Metabolic Engineering Strategies: Integrating Systems and Synthetic Biology with Metabolic Engineering. Trends Biotechnol. 2019, 37, 817–837. [Google Scholar] [CrossRef]
- Qi, L.; Larson, M.H.; Gilbert, L.A.; Doudna, J.A.; Weissman, J.S.; Arkin, A.P.; Lim, W.A. Repurposing CRISPR as an RNA-Guided Platform for Sequence-Specific Control of Gene Expression. Cell 2013, 152, 1173–1183. [Google Scholar] [CrossRef] [Green Version]
- Mccarty, N.S.; Graham, A.E.; Studená, L.; Ledesma-Amaro, R. Multiplexed CRISPR technologies for gene editing and transcriptional regulation. Nat. Commun. 2020, 11, 1–13. [Google Scholar] [CrossRef]
- Ro, D.-K.; Paradise, E.M.; Ouellet, M.; Fisher, K.J.; Newman, K.L.; Ndungu, J.M.; Ho, K.A.; Eachus, R.A.; Ham, T.S.; Kirby, J.; et al. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nat. Cell Biol. 2006, 440, 940–943. [Google Scholar] [CrossRef]
- Martin, V.J.J.; Pitera, D.J.; Withers, S.T.; Newman, J.D.; Keasling, J.D. Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat. Biotechnol. 2003, 21, 796–802. [Google Scholar] [CrossRef] [PubMed]
- Ko, Y.-S.; Kim, J.W.; Lee, J.A.; Han, T.; Kim, G.B.; Park, J.E.; Lee, S.Y. Tools and strategies of systems metabolic engineering for the development of microbial cell factories for chemical production. Chem. Soc. Rev. 2020, 49, 4615–4636. [Google Scholar] [CrossRef] [PubMed]
- Pyne, M.E.; Bruder, M.; Moo-Young, M.; Chung, D.A.; Chou, C.P. Technical guide for genetic advancement of underdeveloped and intractable Clostridium. Biotechnol. Adv. 2014, 32, 623–641. [Google Scholar] [CrossRef] [PubMed]
- Molitor, B.; Kirchner, K.; Henrich, A.W.; Schmitz, S.; Rosenbaum, M.A. Expanding the molecular toolkit for the homoacetogen Clostridium ljungdahlii. Sci. Rep. 2016, 6, 31518. [Google Scholar] [CrossRef] [PubMed]
- Westphal, L.; Wiechmann, A.; Baker, J.; Minton, N.P.; Müller, V. The Rnf Complex Is an Energy-Coupled Transhydrogenase Essential to Reversibly Link Cellular NADH and Ferredoxin Pools in the Acetogen Acetobacterium woodii. J. Bacteriol. 2018, 200. [Google Scholar] [CrossRef] [Green Version]
- Schoelmerich, M.C.; Katsyv, A.; Sung, W.; Mijic, V.; Wiechmann, A.; Kottenhahn, P.; Baker, J.; Minton, N.P.; Müller, V. Regulation of lactate metabolism in the acetogenic bacterium Acetobacterium woodii. Environ. Microbiol. 2018, 20, 4587–4595. [Google Scholar] [CrossRef]
- Wiechmann, A.; Ciurus, S.; Oswald, F.; Seiler, V.N.; Müller, V. It does not always take two to tango: “Syntrophy” via hydrogen cycling in one bacterial cell. Isme J. 2020, 14, 1561–1570. [Google Scholar] [CrossRef]
- Liew, F.; Henstra, A.M.; Köpke, M.; Winzer, K.; Simpson, S.D.; Minton, N.P. Metabolic engineering of Clostridium autoethanogenum for selective alcohol production. Metab. Eng. 2017, 40, 104–114. [Google Scholar] [CrossRef]
- Tremblay, P.-L.; Zhang, T.; Dar, S.A.; Leang, C.; Lovley, D.R. The Rnf Complex of Clostridium ljungdahlii Is a Proton-Translocating Ferredoxin:NAD+ Oxidoreductase Essential for Autotrophic Growth. mBio 2012, 4, e00406-12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leang, C.; Ueki, T.; Nevin, K.P.; Lovley, D.R. A Genetic System for Clostridium ljungdahlii: A Chassis for Autotrophic Production of Biocommodities and a Model Homoacetogen. Appl. Environ. Microbiol. 2012, 79, 1102–1109. [Google Scholar] [CrossRef] [Green Version]
- Ueki, T.; Nevin, K.P.; Woodard, T.L.; Lovley, D.R. Converting Carbon Dioxide to Butyrate with an Engineered Strain of Clostridium ljungdahlii. mBio 2014, 5, e01636-14. [Google Scholar] [CrossRef] [Green Version]
- Huang, H.; Chai, C.; Yang, S.; Jiang, W.; Gu, Y. Phage serine integrase-mediated genome engineering for efficient expression of chemical biosynthetic pathway in gas-fermenting Clostridium ljungdahlii. Metab. Eng. 2019, 52, 293–302. [Google Scholar] [CrossRef] [PubMed]
- Kita, A.; Iwasaki, Y.; Sakai, S.; Okuto, S.; Takaoka, K.; Suzuki, T.; Yano, S.; Sawayama, S.; Tajima, T.; Kato, J.; et al. Development of genetic transformation and heterologous expression system in carboxydotrophic thermophilic acetogen Moorella thermoacetica. J. Biosci. Bioeng. 2013, 115, 347–352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Basen, M.; Geiger, I.; Henke, L.; Müller, V. A Genetic System for the Thermophilic Acetogenic Bacterium Thermoanaerobacter kivui. Appl. Environ. Microbiol. 2017, 84, e02210-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jain, S.; Dietrich, H.M.; Müller, V.; Basen, M. Formate Is Required for Growth of the Thermophilic Acetogenic Bacterium Thermoanaerobacter kivui Lacking Hydrogen-Dependent Carbon Dioxide Reductase (HDCR). Front. Microbiol. 2020, 11, 59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mock, J.; Zheng, Y.; Mueller, A.P.; Ly, S.; Tran, L.; Segovia, S.; Nagaraju, S.; Köpke, M.; Dürre, P.; Thauer, R.K. Energy Conservation Associated with Ethanol Formation from H2 and CO2 in Clostridium autoethanogenum Involving Electron Bifurcation. J. Bacteriol. 2015, 197, 2965–2980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marcellin, E.; Behrendorff, J.B.; Nagaraju, S.; DeTissera, S.; Segovia, S.; Palfreyman, R.W.; Daniell, J.; Licona-Cassani, C.; Quek, L.-E.; Speight, R.; et al. Low carbon fuels and commodity chemicals from waste gases—Systematic approach to understand energy metabolism in a model acetogen. Green Chem. 2016, 18, 3020–3028. [Google Scholar] [CrossRef] [Green Version]
- Liew, F.; Henstra, A.M.; Winzer, K.; Köpke, M.; Simpson, S.D.; Minton, N.P. Insights into CO2 Fixation Pathway of Clostridium autoethanogenum by Targeted Mutagenesis. mBio 2016, 7, e00427-16. [Google Scholar] [CrossRef] [Green Version]
- Bengelsdorf, F.R.; Poehlein, A.; Linder, S.; Erz, C.; Hummel, T.; Hoffmeister, S.; Daniel, R.; Dürre, P. Industrial Acetogenic Biocatalysts: A Comparative Metabolic and Genomic Analysis. Front. Microbiol. 2016, 7, 1036. [Google Scholar] [CrossRef] [Green Version]
- Nagaraju, S.; Davies, N.K.; Walker, D.J.F.; Köpke, M.; Simpson, S.D. Genome editing of Clostridium autoethanogenum using CRISPR/Cas9. Biotechnol. Biofuels 2016, 9, 219. [Google Scholar] [CrossRef] [Green Version]
- Huang, H.; Chai, C.; Li, N.; Rowe, P.; Minton, N.P.; Yang, S.; Jiang, W.; Gu, Y. CRISPR/Cas9-Based Efficient Genome Editing in Clostridium ljungdahlii, an Autotrophic Gas-Fermenting Bacterium. ACS Synth. Biol. 2016, 5, 1355–1361. [Google Scholar] [CrossRef]
- Zhao, R.; Liu, Y.; Zhang, H.; Chai, C.; Wang, J.; Jiang, W.; Gu, Y. CRISPR-Cas12a-Mediated Gene Deletion and Regulation in Clostridium ljungdahlii and Its Application in Carbon Flux Redirection in Synthesis Gas Fermentation. ACS Synth. Biol. 2019, 8, 2270–2279. [Google Scholar] [CrossRef]
- Xia, P.-F.; Casini, I.; Schulz, S.; Klask, C.-M.; Angenent, L.T.; Molitor, B. Reprogramming Acetogenic Bacteria with CRISPR-Targeted Base Editing via Deamination. ACS Synth. Biol. 2020, 9, 2162–2171. [Google Scholar] [CrossRef]
- Shin, J.; Kang, S.; Song, Y.; Jin, S.; Lee, J.S.; Lee, J.-K.; Kim, D.R.; Kim, S.C.; Cho, S.; Cho, B.-K. Genome Engineering of Eubacterium limosum Using Expanded Genetic Tools and the CRISPR-Cas9 System. ACS Synth. Biol. 2019, 8, 2059–2068. [Google Scholar] [CrossRef] [PubMed]
- Jeong, J.; Kim, J.-Y.; Park, B.; Choi, I.-G.; Chang, I.S. Genetic engineering system for syngas-utilizing acetogen, Eubacterium limosum KIST612. Bioresour. Technol. Rep. 2020, 11, 100452. [Google Scholar] [CrossRef]
- Köpke, M.; Held, C.; Hujer, S.; Liesegang, H.; Wiezer, A.; Wollherr, A.; Ehrenreich, A.; Liebl, W.; Gottschalk, G.; Dürre, P. Clostridium ljungdahlii represents a microbial production platform based on syngas. Proc. Natl. Acad. Sci. USA 2010, 107, 13087–13092. [Google Scholar] [CrossRef] [Green Version]
- Becker, F.U.; Grund, G.; Orschel, M.; Doderer, K.; Löhden, G.; Brand, G. Cells and Method for Producing Acetone. U.S. Patent 20120101304 A1, 26 April 2012. [Google Scholar]
- Han, S.; Gao, X.-Y.; Ying, H.; Zhou, C.C. NADH gene manipulation for advancing bioelectricity in Clostridium ljungdahlii microbial fuel cells. Green Chem. 2016, 18, 2473–2478. [Google Scholar] [CrossRef]
- Yang, G.; Jia, D.; Jin, L.; Jiang, Y.; Wang, Y.; Jiang, W.; Gu, Y. Rapid Generation of Universal Synthetic Promoters for Controlled Gene Expression in Both Gas-Fermenting and Saccharolytic Clostridium Species. ACS Synth. Biol. 2017, 6, 1672–1678. [Google Scholar] [CrossRef] [PubMed]
- Straub, M.; Demler, M.; Weuster-Botz, D.; Dürre, P. Selective enhancement of autotrophic acetate production with genetically modified Acetobacterium woodii. J. Biotechnol. 2014, 178, 67–72. [Google Scholar] [CrossRef] [PubMed]
- Hoffmeister, S.; Gerdom, M.; Bengelsdorf, F.R.; Linder, S.; Flüchter, S.; Öztürk, H.; Blümke, W.; May, A.; Fischer, R.-J.; Bahl, H.; et al. Acetone production with metabolically engineered strains of Acetobacterium woodii. Metab. Eng. 2016, 36, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Lee, J.S.; Shin, J.; Lee, G.M.; Jin, S.; Kang, S.; Lee, J.-K.; Kim, D.R.; Lee, E.Y.; Kim, S.C.; et al. Functional cooperation of the glycine synthase-reductase and Wood–Ljungdahl pathways for autotrophic growth of Clostridium drakei. Proc. Natl. Acad. Sci. USA 2020, 117, 7516–7523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, S.; Song, Y.; Jin, S.; Shin, J.; Bae, J.; Kim, D.R.; Lee, J.-K.; Kim, S.C.; Cho, S.; Cho, B.-K. Adaptive Laboratory Evolution of Eubacterium limosum ATCC 8486 on Carbon Monoxide. Front. Microbiol. 2020, 11, 402. [Google Scholar] [CrossRef] [PubMed]
- Heap, J.T.; Pennington, O.J.; Cartman, S.T.; Minton, N.P. A modular system for Clostridium shuttle plasmids. J. Microbiol. Methods 2009, 78, 79–85. [Google Scholar] [CrossRef] [PubMed]
- Rahayu, F.; Kawai, Y.; Iwasaki, Y.; Yoshida, K.; Kita, A.; Tajima, T.; Kato, J.; Murakami, K.; Hoshino, T.; Nakashimada, Y. Thermophilic ethanol fermentation from lignocellulose hydrolysate by genetically engineered Moorella thermoacetica. Bioresour. Technol. 2017, 245, 1393–1399. [Google Scholar] [CrossRef] [PubMed]
- Diner, B.A.; Fan, J.; Scotcher, M.C.; Wells, D.H.; Whited, G.M. Synthesis of Heterologous Mevalonic Acid Pathway Enzymes in Clostridium ljungdahlii for the Conversion of Fructose and of Syngas to Mevalonate and Isoprene. Appl. Environ. Microbiol. 2017, 84, e01723-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koepke, M.; Chen, W.Y. Recombinant Microorganisms and Uses Therefor. U.S. Patent 20130323806 A1, 5 December 2013. [Google Scholar]
- Nagaraju, S.; Al-Sinawi, B.; Tissera, S.D.; Koepke, M. Recombinant Microorganisms and Methods of use Thereof. U.S. Patent 20150210987 A1, 30 July 2015. [Google Scholar]
- Köpke, M.; Gerth, M.L.; Maddock, D.J.; Mueller, A.P.; Liew, F.; Simpson, S.D.; Patrick, W.M. Reconstruction of an Acetogenic 2,3-Butanediol Pathway Involving a Novel NADPH-Dependent Primary-Secondary Alcohol Dehydrogenase. Appl. Environ. Microbiol. 2014, 80, 3394–3403. [Google Scholar] [CrossRef] [Green Version]
- Bron, S.; Luxen, E. Segregational instability of pUB110-derived recombinant plasmids in Bacillus subtilis. Plasmid 1985, 14, 235–244. [Google Scholar] [CrossRef]
- Friehs, K. Plasmid Copy Number and Plasmid Stability; Springer Science and Business Media LLC: Berlin, Germany, 2003; Volume 86, pp. 47–82. [Google Scholar]
- Borkowski, O.; Ceroni, F.; Stan, G.-B.; Ellis, T. Overloaded and stressed: Whole-cell considerations for bacterial synthetic biology. Curr. Opin. Microbiol. 2016, 33, 123–130. [Google Scholar] [CrossRef]
- Strätz, M.; Gottschalk, G.; Dürre, P. Transfer and expression of the tetracyclin resistance transposon Tn925 in Acetobacterium woodii. Fems Microbiol. Lett. 1990, 68, 171–176. [Google Scholar] [CrossRef]
- Philipps, G.; De Vries, S.; Jennewein, S. Development of a metabolic pathway transfer and genomic integration system for the syngas-fermenting bacterium Clostridium ljungdahlii. Biotechnol. Biofuels 2019, 12, 112. [Google Scholar] [CrossRef]
- Karberg, M.; Guo, H.; Zhong, J.; Coon, R.; Perutka, J.; Lambowitz, A.M. Group II introns as controllable gene targeting vectors for genetic manipulation of bacteria. Nat. Biotechnol. 2001, 19, 1162–1167. [Google Scholar] [CrossRef]
- Lambowitz, A.M.; Zimmerly, S. Group II Introns: Mobile Ribozymes that Invade DNA. Cold Spring Harb. Perspect. Biol. 2010, 3, a003616. [Google Scholar] [CrossRef] [PubMed]
- Heap, J.T.; Pennington, O.J.; Cartman, S.T.; Carter, G.P.; Minton, N.P. The ClosTron: A universal gene knock-out system for the genus Clostridium. J. Microbiol. Methods 2007, 70, 452–464. [Google Scholar] [CrossRef] [PubMed]
- Heap, J.T.; Kuehne, S.A.; Ehsaan, M.; Cartman, S.T.; Cooksley, C.M.; Scott, J.C.; Minton, N.P. The ClosTron: Mutagenesis in Clostridium refined and streamlined. J. Microbiol. Methods 2010, 80, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Kuehne, S.A.; Minton, N.P. ClosTron-mediated engineering of Clostridium. Bioengineered 2012, 3, 247–254. [Google Scholar] [CrossRef] [Green Version]
- Plante, I.; Cousineau, B. Restriction for gene insertion within the Lactococcus lactis Ll.LtrB group II intron. RNA 2006, 12, 1980–1992. [Google Scholar] [CrossRef] [Green Version]
- Lampe, D.J.; Churchill, M.E.; Robertson, H.M. A purified mariner transposase is sufficient to mediate transposition in vitro. Embo J. 1996, 15, 5470–5479. [Google Scholar] [CrossRef]
- Zhang, Y.; Grosse-Honebrink, A.; Minton, N.P. A Universal Mariner Transposon System for Forward Genetic Studies in the Genus Clostridium. PLoS ONE 2015, 10, e0122411. [Google Scholar] [CrossRef] [Green Version]
- Fu, J.; Wenzel, S.C.; Perlova, O.; Wang, J.; Gross, F.; Tang, Z.; Yin, Y.; Stewart, A.F.; Müller, R.; Zhang, Y. Efficient transfer of two large secondary metabolite pathway gene clusters into heterologous hosts by transposition. Nucl. Acids Res. 2008, 36, e113. [Google Scholar] [CrossRef] [Green Version]
- Heap, J.T.; Ehsaan, M.; Cooksley, C.M.; Ng, Y.-K.; Cartman, S.T.; Winzer, K.; Minton, N.P. Integration of DNA into bacterial chromosomes from plasmids without a counter-selection marker. Nucl. Acids Res. 2012, 40, e59. [Google Scholar] [CrossRef]
- Minton, N.P.; Ehsaan, M.; Humphreys, C.M.; Little, G.T.; Baker, J.; Henstra, A.M.; Liew, F.; Kelly, M.L.; Sheng, L.; Schwarz, K.; et al. A roadmap for gene system development in Clostridium. Anaerobe 2016, 41, 104–112. [Google Scholar] [CrossRef]
- Jiang, W.; Bikard, D.; Cox, D.; Zhang, F.; Marraffini, L.A. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat. Biotechnol. 2013, 31, 233–239. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.D.; Lander, E.S.; Zhang, F. Development and Applications of CRISPR-Cas9 for Genome Engineering. Cell 2014, 157, 1262–1278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.; et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Makarova, K.S.; Wolf, Y.I.; Alkhnbashi, O.S.; Costa, F.; Shah, S.A.; Saunders, S.J.; Barrangou, R.; Brouns, S.J.; Charpentier, E.; Haft, D.H.; et al. An updated evolutionary classification of CRISPR–Cas systems. Nat. Rev. Genet. 2015, 13, 722–736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cañadas, I.C.; Groothuis, D.; Zygouropoulou, M.; Rodrigues, R.; Minton, N.P. RiboCas: A Universal CRISPR-Based Editing Tool for Clostridium. ACS Synth. Biol. 2019, 8, 1379–1390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seys, F.M.; Rowe, P.; Bolt, E.L.; Humphreys, C.M.; Minton, N.P. A Gold Standard, CRISPR/Cas9-Based Complementation Strategy Reliant on 24 Nucleotide Bookmark Sequences. Genes 2020, 11, 458. [Google Scholar] [CrossRef] [Green Version]
- Zetsche, B.; Gootenberg, J.S.; Abudayyeh, O.O.; Slaymaker, I.M.; Makarova, K.S.; Essletzbichler, P.; Volz, S.E.; Joung, J.; Van Der Oost, J.; Regev, A.; et al. Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Cell 2015, 163, 759–771. [Google Scholar] [CrossRef] [Green Version]
- Ao, X.; Yao, Y.; Li, T.; Yang, T.-T.; Dong, X.; Zheng, Z.-T.; Chen, G.-Q.; Wu, Q.; Guo, Y. A Multiplex Genome Editing Method for Escherichia coli Based on CRISPR-Cas12a. Front. Microbiol. 2018, 9, 2307. [Google Scholar] [CrossRef]
- Adiego-Pérez, B.; Randazzo, P.; Daran, J.M.; Verwaal, R.; Roubos, J.A.; Daran-Lapujade, P.; Van Der Oost, J. Multiplex genome editing of microorganisms using CRISPR-Cas. Fems Microbiol. Lett. 2019, 366, 366. [Google Scholar] [CrossRef]
- Zetsche, B.; Heidenreich, M.; Mohanraju, P.; Fedorova, I.; Kneppers, J.; DeGennaro, E.M.; Winblad, N.; Choudhury, S.R.; Abudayyeh, O.O.; Gootenberg, J.S.; et al. Multiplex gene editing by CRISPR–Cpf1 using a single crRNA array. Nat. Biotechnol. 2016, 35, 31–34. [Google Scholar] [CrossRef]
- Koepke, M.; Liew, F. Genetically Engineered Bacterium with Altered Carbon Monoxide Dehydrogenase (codh) Activity. U.S. Patent 20160040193 A1, 11 February 2016. [Google Scholar]
- Figueroa, I.A.; Barnum, T.P.; Somasekhar, P.Y.; Carlström, C.I.; Engelbrektson, A.L.; Coates, J.D. Metagenomics-guided analysis of microbial chemolithoautotrophic phosphite oxidation yields evidence of a seventh natural CO2 fixation pathway. Proc. Natl. Acad. Sci. USA 2017, 115, E92–E101. [Google Scholar] [CrossRef] [Green Version]
- Bertsch, J.; Müller, V. Bioenergetic constraints for conversion of syngas to biofuels in acetogenic bacteria. Biotechnol. Biofuels 2015, 8, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Richter, H.; Molitor, B.; Wei, H.; Chen, W.; Aristilde, L.; Angenent, L.T. Ethanol production in syngas-fermenting Clostridium ljungdahlii is controlled by thermodynamics rather than by enzyme expression. Energ. Environ. Sci. 2016, 9, 2392–2399. [Google Scholar] [CrossRef]
- Banerjee, A.; Leang, C.; Ueki, T.; Nevin, K.P.; Lovley, D.R. Lactose-Inducible System for Metabolic Engineering of Clostridium ljungdahlii. Appl. Environ. Microbiol. 2014, 80, 2410–2416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mahamkali, V.; Valgepea, K.; Lemgruber, R.D.S.P.; Plan, M.; Tappel, R.; Köpke, M.; Simpson, S.D.; Nielsen, L.K.; Marcellin, E. Redox controls metabolic robustness in the gas-fermenting acetogen Clostridium autoethanogenum. Proc. Natl. Acad. Sci. USA 2020, 117, 13168–13175. [Google Scholar] [CrossRef]
- Simpson, S.D.; Koepke, M.; Liew, F. Recombinant Microorganisms and Methods of use Thereof. U.S. Patent 20110256600 A1, 20 October 2011. [Google Scholar]
- Mueller, A.P.; Koepke, M.; Nagaraju, S. Recombinant microorganisms and uses therefor. U.S. Patent 20130330809 A1, 12 December 2013. [Google Scholar]
- Liao, C.; Seo, S.-O.; Celik, V.; Liu, H.; Kong, W.; Wang, Y.; Blaschek, H.; Jin, Y.-S.; Lu, T. Integrated, systems metabolic picture of acetone-butanol-ethanol fermentation by Clostridium acetobutylicum. Proc. Natl. Acad. Sci. USA 2015, 112, 8505–8510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, N.-P.-T.; Raynaud, C.; Meynial-Salles, I.; Soucaille, P. Reviving the Weizmann process for commercial n-butanol production. Nat. Commun. 2018, 9, 3682. [Google Scholar] [CrossRef]
- Koepke, M.; Simpson, S.D.; Liew, F.; Chen, W. Fermentation Process for Producing Isopropanol Usinga Recombinant Microorganism. U.S. Patent 20120252083 A1, 4 October 2012. [Google Scholar]
- Koepke, M.; Liew, F. Recombinant Microorganismand Methods of Production Thereof. U.S. Patent 20110236941 A1, 29 September 2011. [Google Scholar]
- Chen, W.Y.; Liew, F.; Koepke, M. Recombinant Microorganisms and Uses Therefor. U.S. Patent 20130323820 A1, 5 December 2013. [Google Scholar]
- Beeby, M.; Cho, M.; Stubbe, J.; Jensen, G.J. Growth and Localization of Polyhydroxybutyrate Granules in Ralstonia eutropha. J. Bacteriol. 2011, 194, 1092–1099. [Google Scholar] [CrossRef] [Green Version]
- Bhagowati, P.; Pradhan, S.; Dash, H.R.; Das, S. Production, optimization and characterization of polyhydroxybutyrate, a biodegradable plastic by Bacillus spp. Biosci. Biotechnol. Biochem. 2015, 79, 1454–1463. [Google Scholar] [CrossRef] [Green Version]
- Lemgruber, R.D.S.P.; Valgepea, K.; Tappel, R.; Behrendorff, J.B.; Palfreyman, R.W.; Plan, M.; Hodson, M.P.; Simpson, S.D.; Nielsen, L.K.; Köpke, M.; et al. Systems-level engineering and characterisation of Clostridium autoethanogenum through heterologous production of poly-3-hydroxybutyrate (PHB). Metab. Eng. 2019, 53, 14–23. [Google Scholar] [CrossRef]
- Beck, Z.Q.; Cervin, M.A.; Chotani, G.K.; Diner, B.A.; Fan, J. Recombinant Anaerobc Acetogenic Bacteria for Production of Isoprene and/or Industrial Bo-Products Using Synthesis Gas. U.S. Patent 20140234926 A1, 21 August 2014. [Google Scholar]
- Annan, F.J.; Al-Sinawi, B.; Humphreys, C.M.; Norman, R.; Winzer, K.; Köpke, M.; Simpson, S.D.; Minton, N.P.; Henstra, A.M. Engineering of vitamin prototrophy in Clostridium ljungdahlii and Clostridium autoethanogenum. Appl. Microbiol. Biotechnol. 2019, 103, 4633–4648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hossain, G.S.; Saini, M.; Miyake, R.; Chang, M.W.; Chang, M.W. Genetic Biosensor Design for Natural Product Biosynthesis in Microorganisms. Trends Biotechnol. 2020, 38, 797–810. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Liu, J.; Chen, X.; Zheng, H.; Li, F. RNA-seq-based comparative transcriptome analysis of the syngas-utilizing bacterium Clostridium ljungdahlii DSM 13528 grown autotrophically and heterotrophically. Mol. Biosyst. 2013, 9, 2775–2784. [Google Scholar] [CrossRef] [PubMed]
- Utturkar, S.M.; Klingeman, D.M.; Bruno-Barcena, J.M.; Chinn, M.S.; Grunden, A.M.; Köpke, M.; Brown, S.D. Sequence data for Clostridium autoethanogenum using three generations of sequencing technologies. Sci. Data 2015, 2, 150014. [Google Scholar] [CrossRef] [PubMed]
- Whitham, J.M.; Tirado-Acevedo, O.; Chinn, M.S.; Pawlak, J.J.; Grunden, A.M. Metabolic Response of Clostridium ljungdahlii to Oxygen Exposure. Appl. Environ. Microbiol. 2015, 81, 8379–8391. [Google Scholar] [CrossRef] [Green Version]
- Song, Y.; Shin, J.; Jeong, Y.; Jin, S.; Lee, J.-K.; Kim, D.R.; Kim, S.C.; Cho, S.; Cho, B.-K. Determination of the Genome and Primary Transcriptome of Syngas Fermenting Eubacterium limosum ATCC 8486. Sci. Rep. 2017, 7, 13694. [Google Scholar] [CrossRef] [PubMed]
- Kremp, F.; Poehlein, A.; Daniel, R.; Müller, V. Methanol metabolism in the acetogenic bacterium Acetobacterium woodii. Environ. Microbiol. 2018, 20, 4369–4384. [Google Scholar] [CrossRef]
- Shin, J.; Song, Y.; Jin, S.; Lee, J.-K.; Kim, D.R.; Kim, S.C.; Cho, S.; Cho, B.-K. Genome-scale analysis of Acetobacterium bakii reveals the cold adaptation of psychrotolerant acetogens by post-transcriptional regulation. RNA 2018, 24, 1839–1855. [Google Scholar] [CrossRef] [Green Version]
- Al-Bassam, M.M.; Kim, J.-N.; Zaramela, L.S.; Kellman, B.P.; Zuniga, C.; Wozniak, J.M.; Gonzalez, D.J.; Zengler, K. Optimization of carbon and energy utilization through differential translational efficiency. Nat. Commun. 2018, 9, 4474. [Google Scholar] [CrossRef] [Green Version]
- Song, Y.; Shin, J.; Jin, S.; Lee, J.-K.; Kim, D.R.; Kim, S.C.; Cho, S.; Cho, B.-K. Genome-scale analysis of syngas fermenting acetogenic bacteria reveals the translational regulation for its autotrophic growth. BMC Genom. 2018, 19, 837. [Google Scholar] [CrossRef]
- Valgepea, K.; Lemgruber, R.D.S.P.; Meaghan, K.; Palfreyman, R.W.; Abdalla, T.; Heijstra, B.D.; Behrendorff, J.B.Y.H.; Tappel, R.; Köpke, M.; Simpson, S.D.; et al. Maintenance of ATP Homeostasis Triggers Metabolic Shifts in Gas-Fermenting Acetogens. Cell Syst. 2017, 4, 505–515.e5. [Google Scholar] [CrossRef] [Green Version]
- Greene, J.; Daniell, J.; Köpke, M.; Broadbelt, L.J.; Tyo, K.E. Kinetic ensemble model of gas fermenting Clostridium autoethanogenum for improved ethanol production. Biochem. Eng. J. 2019, 148, 46–56. [Google Scholar] [CrossRef]
- Liu, J.K.; Lloyd, C.; Al-Bassam, M.M.; Ebrahim, A.; Kim, J.-N.; Olson, C.A.; Aksenov, A.; Dorrestein, P.; Zengler, K. Predicting proteome allocation, overflow metabolism, and metal requirements in a model acetogen. PLoS Comput. Biol. 2019, 15, e1006848. [Google Scholar] [CrossRef]
- Zhu, H.-F.; Liu, Z.-Y.; Zhou, X.; Yi, J.-H.; Lun, Z.-M.; Wang, S.-N.; Tang, W.-Z.; Li, F.-L. Energy Conservation and Carbon Flux Distribution during Fermentation of CO or H2/CO2 by Clostridium ljungdahlii. Front. Microbiol. 2020, 11, 416. [Google Scholar] [CrossRef]
- Heffernan, J.K.; Valgepea, K.; Lemgruber, R.D.S.P.; Casini, I.; Plan, M.; Tappel, R.; Simpson, S.D.; Köpke, M.; Nielsen, L.K.; Marcellin, E. Enhancing CO2-Valorization Using Clostridium autoethanogenum for Sustainable Fuel and Chemicals Production. Front. Bioeng. Biotechnol. 2020, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagarajan, H.; Sahin, M.; Nogales, J.; Latif, H.; Lovley, D.R.; Ebrahim, A.; Zengler, K. Characterizing acetogenic metabolism using a genome-scale metabolic reconstruction of Clostridium ljungdahlii. Microb. Cell Fact. 2013, 12, 118. [Google Scholar] [CrossRef] [Green Version]
- Brown, S.D.; Nagaraju, S.; Utturkar, S.M.; De Tissera, S.; Segovia, S.; Mitchell, W.; Land, M.L.; Dassanayake, A.; Köpke, M. Comparison of single-molecule sequencing and hybrid approaches for finishing the genome of Clostridium autoethanogenum and analysis of CRISPR systems in industrial relevant Clostridia. Biotechnol. Biofuels 2014, 7, 40. [Google Scholar] [CrossRef]
- Poehlein, A.; Cebulla, M.; Ilg, M.M.; Bengelsdorf, F.R.; Schiel-Bengelsdorf, B.; Whited, G.; Andreesen, J.R.; Gottschalk, G.; Daniel, R.; Dürre, P. The Complete Genome Sequence of Clostridium aceticum: A Missing Link between Rnf- and Cytochrome-Containing Autotrophic Acetogens. mBio 2015, 6, e01168-15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Humphreys, C.M.; McLean, S.; Schatschneider, S.; Millat, T.; Henstra, A.M.; Annan, F.J.; Breitkopf, F.; Pander, B.; Piatek, P.; Rowe, P.; et al. Whole genome sequence and manual annotation of Clostridium autoethanogenum, an industrially relevant bacterium. BMC Genom. 2015, 16, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Shin, J.; Song, Y.; Jeong, Y.; Cho, B.-K. Analysis of the Core Genome and Pan-Genome of Autotrophic Acetogenic Bacteria. Front. Microbiol. 2016, 7, 1531. [Google Scholar] [CrossRef] [PubMed]
- Aklujkar, M.; Leang, C.; Shrestha, P.M.; Shrestha, M.; Lovley, D.R. Transcriptomic profiles of Clostridium ljungdahlii during lithotrophic growth with syngas or H2 and CO2 compared to organotrophic growth with fructose. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Valgepea, K.; Lemgruber, R.D.S.P.; Abdalla, T.; Binos, S.; Takemori, N.; Takemori, A.; Tanaka, Y.; Tappel, R.; Köpke, M.; Simpson, S.D.; et al. H2 drives metabolic rearrangements in gas-fermenting Clostridium autoethanogenum. Biotechnol. Biofuels 2018, 11, 1–15. [Google Scholar] [CrossRef]
- Esposito, A.; Tamburini, S.; Triboli, L.; Ambrosino, L.; Chiusano, M.L.; Jousson, O. Insights into the genome structure of four acetogenic bacteria with specific reference to the Wood–Ljungdahl pathway. Microbiol. 2019, 8, e938. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Lee, J.W.; Chae, C.G.; Kwon, S.J.; Kim, Y.J.; Lee, J.-H.; Lee, H.S. Domestication of the novel alcohologenic acetogen Clostridium sp. AWRP: From isolation to characterization for syngas fermentation. Biotechnol. Biofuels 2019, 12, 1–14. [Google Scholar] [CrossRef]
- Wang, S.E.; Brooks, A.E.; Cann, B.; Simoes-Barbosa, A. The fluorescent protein iLOV outperforms eGFP as a reporter gene in the microaerophilic protozoan Trichomonas vaginalis. Mol. Biochem. Parasitol. 2017, 216, 1–4. [Google Scholar] [CrossRef]
- Zhang, C.; Xing, X.-H.; Lou, K. Rapid detection of a gfp-marked Enterobacter aerogenes under anaerobic conditions by aerobic fluorescence recovery. FEMS Microbiol. Lett. 2005, 249, 211–218. [Google Scholar] [CrossRef] [Green Version]
- Buckley, A.M.; Petersen, J.; Roe, A.J.; Douce, G.R.; Christie, J.M. LOV-based reporters for fluorescence imaging. Curr. Opin. Chem. Biol. 2015, 27, 39–45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buckley, A.M.; Jukes, C.; Candlish, D.; Irvine, J.J.; Spencer, J.; Fagan, R.P.; Roe, A.J.; Christie, J.M.; Fairweather, N.F.; Douce, G.R. Lighting Up Clostridium Difficile: Reporting Gene Expression Using Fluorescent Lov Domains. Sci. Rep. 2016, 6, 23463. [Google Scholar] [CrossRef] [Green Version]
- Streett, H.E.; Kalis, K.M.; Papoutsakis, E.T. A Strongly Fluorescing Anaerobic Reporter and Protein-Tagging System for Clostridium Organisms Based on the Fluorescence-Activating and Absorption-Shifting Tag Protein (FAST). Appl. Environ. Microbiol. 2019, 85. [Google Scholar] [CrossRef] [Green Version]
- Drepper, T.; Huber, R.; Heck, A.; Circolone, F.; Hillmer, A.-K.; Büchs, J.; Jaeger, K.-E. Flavin Mononucleotide-Based Fluorescent Reporter Proteins Outperform Green Fluorescent Protein-Like Proteins as Quantitative In Vivo Real-Time Reporters. Appl. Environ. Microbiol. 2010, 76, 5990–5994. [Google Scholar] [CrossRef] [Green Version]
- Yeom, S.-J.; Kim, M.; Kwon, K.K.; Fu, Y.; Rha, E.; Park, S.-H.; Lee, H.; Kim, H.; Lee, D.-H.; Kim, D.-M.; et al. A synthetic microbial biosensor for high-throughput screening of lactam biocatalysts. Nat. Commun. 2018, 9, 5053. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raman, S.; Rogers, J.K.; Taylor, N.D.; Church, G.M. Evolution-guided optimization of biosynthetic pathways. Proc. Natl. Acad. Sci. USA 2014, 111, 17803–17808. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, D.; Kim, W.J.; Yoo, S.M.; Choi, J.H.; Ha, S.H.; Lee, M.H.; Lee, S.Y. Repurposing type III polyketide synthase as a malonyl-CoA biosensor for metabolic engineering in bacteria. Proc. Natl. Acad. Sci. USA 2018, 115, 9835–9844. [Google Scholar] [CrossRef] [Green Version]
- Bikard, D.; Jiang, W.; Samai, P.; Hochschild, A.; Zhang, F.; Marraffini, L.A. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res. 2013, 41, 7429–7437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Konermann, S.; Brigham, M.D.; Trevino, A.E.; Joung, J.; Abudayyeh, O.O.; Barcena, C.; Hsu, P.D.; Habib, N.; Gootenberg, J.S.; Nishimasu, H.; et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nat. Cell Biol. 2014, 517, 583–588. [Google Scholar] [CrossRef] [Green Version]
- Dong, C.; Fontana, J.; Patel, A.; Carothers, J.M.; Zalatan, J.G. Synthetic CRISPR-Cas gene activators for transcriptional reprogramming in bacteria. Nat. Commun. 2018, 9, 2489. [Google Scholar] [CrossRef] [Green Version]
- Cleto, S.; Jensen, J.V.; Wendisch, V.F.; Lu, T.K. Corynebacterium glutamicum Metabolic Engineering with CRISPR Interference (CRISPRi). ACS Synth. Biol. 2016, 5, 375–385. [Google Scholar] [CrossRef]
- Wen, Z.; Minton, N.P.; Zhang, Y.; Li, Q.; Liu, J.; Jiang, Y.; Yang, S. Enhanced solvent production by metabolic engineering of a twin-clostridial consortium. Metab. Eng. 2017, 39, 38–48. [Google Scholar] [CrossRef]
- Westbrook, A.W.; Moo-Young, M.; Chou, C.P. Development of a CRISPR-Cas9 Tool Kit for Comprehensive Engineering of Bacillus subtilis. Appl. Environ. Microbiol. 2016, 82, 4876–4895. [Google Scholar] [CrossRef] [Green Version]
- Wu, M.-Y.; Sung, L.-Y.; Li, H.; Huang, C.-H.; Hu, Y.-C. Combining CRISPR and CRISPRi Systems for Metabolic Engineering of E. coli and 1,4-BDO Biosynthesis. ACS Synth. Biol. 2017, 6, 2350–2361. [Google Scholar] [CrossRef]
- Kim, S.K.; Han, G.H.; Seong, W.; Kim, H.; Kim, S.-W.; Lee, D.-H.; Lee, S.-G. CRISPR interference-guided balancing of a biosynthetic mevalonate pathway increases terpenoid production. Metab. Eng. 2016, 38, 228–240. [Google Scholar] [CrossRef] [PubMed]
- Lv, L.; Ren, Y.-L.; Chen, J.-C.; Wu, Q.; Chen, G.-Q. Application of CRISPRi for prokaryotic metabolic engineering involving multiple genes, a case study: Controllable P(3HB-co-4HB) biosynthesis. Metab. Eng. 2015, 29, 160–168. [Google Scholar] [CrossRef]
- Woolston, B.M.; Emerson, D.F.; Currie, D.H.; Stephanopoulos, G. Rediverting carbon flux in Clostridium ljungdahlii using CRISPR interference (CRISPRi). Metab. Eng. 2018, 48, 243–253. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.H.; Ostrov, N.; Wong, B.G.; Gold, M.A.; Khalil, A.S.; Church, G.M. Functional genomics of the rapidly replicating bacterium Vibrio natriegens by CRISPRi. Nat. Microbiol. 2019, 4, 1105–1113. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Gallay, C.; Kjos, M.; Domenech, A.; Slager, J.; Van Kessel, S.P.; Knoops, K.; Sorg, R.A.; Zhang, J.-R.; Veening, J.-W. High-throughput CRISPRi phenotyping identifies new essential genes in Streptococcus pneumoniae. Mol. Syst. Biol. 2017, 13, 931. [Google Scholar] [CrossRef]
- Peters, J.M.; Colavin, A.; Shi, H.; Czarny, T.L.; Larson, M.H.; Wong, S.; Hawkins, J.S.; Lu, C.H.S.; Koo, B.-M.; Marta, E.; et al. A Comprehensive, CRISPR-based Functional Analysis of Essential Genes in Bacteria. Cell 2016, 165, 1493–1506. [Google Scholar] [CrossRef] [Green Version]
- Wang, T.; Guan, C.; Guo, J.; Liu, B.; Wu, Y.; Xie, Z.; Zhang, C.; Xing, X.-H. Pooled CRISPR interference screening enables genome-scale functional genomics study in bacteria with superior performance. Nat. Commun. 2018, 9, 2475. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, A.A.K.; Voigt, C.A. Multi-input CRISPR / C as genetic circuits that interface host regulatory networks. Mol. Syst. Biol. 2014, 10, 763. [Google Scholar] [CrossRef]
- Cress, B.F.; Jones, J.A.; Kim, D.C.; Leitz, Q.D.; Englaender, J.A.; Collins, S.M.; Linhardt, R.J.; Koffas, M.A.G. Rapid generation of CRISPR/dCas9-regulated, orthogonally repressible hybrid T7-lac promoters for modular, tuneable control of metabolic pathway fluxes in Escherichia coli. Nucleic Acids Res. 2016, 44, 4472–4485. [Google Scholar] [CrossRef] [Green Version]
- Tremblay, P.-L.; Höglund, D.; Koza, A.; Bonde, I.; Zhang, T. Adaptation of the autotrophic acetogen Sporomusa ovata to methanol accelerates the conversion of CO2 to organic products. Sci. Rep. 2015, 5, 16168. [Google Scholar] [CrossRef] [Green Version]



| Plasmid | Gram (+) Replicon | Marker | Gram (−) Replicon | Applicable Species | Ref. |
|---|---|---|---|---|---|
| pIMP1 | pIM13 (repL) | amp/ermC | ColE1 | Clostridium aceticum, Clostridium ljungdahlii | [46,47] |
| pJIR750ai | pIP404 | catP | ColE1 | Acetobacterium woodii, Eubacterium limosum, Clostridium ljungdahlii | [44,50,52,53] |
| pK18mobsacB * | RP4 | catP/ermB | pUC | Moorella thermoacetica | [33,55] |
| pMTL82151 | pBP1 (repA) | catP | ColE1 | Clostridium ljungdahlii, Clostridium autoethanogenum, Eubacterium limosum | [24,30,44,54] |
| pMTL82254 | pBP1 (repA) | ermB | ColE1 | Clostridium ljungdahlii, Clostridium autoethanogenum, Eubacterium limosum | [44,54] |
| pMTL83151 | pCB102 (repH) | catP | ColE1 | Clostridium ljungdahlii, Clostridium autoethanogenum, Eubacterium limosum | [24,28,44,54,56] |
| pMTL83245 | pIM13 (repL) | ermB | ColE1 | Clostridium ljungdahlii, Clostridium autoethanogenum | [54,57] |
| pMTL83353 | pCB102 (repH) | aad9 | ColE1 | Clostridium ljungdahlii, Clostridium autoethanogenum | [54] |
| pMTL84151 | pCD6 (repA) | catP | ColE1 | Clostridium ljungdahlii, Clostridium autoethanogenum, Eubacterium limosum | [28,44,51,54] |
| pMTL84422 | pCD6 (repA) | tetA(P) | p15a | Clostridium ljungdahlii, Clostridium autoethanogenum | [54] |
| pMTL85151 | pIM13 (repL) | catP | ColE1 | Clostridium ljungdahlii, Clostridium autoethanogenum, Eubacterium limosum | [44,54,58] |
| pMTL85241 | pIM13 (repL) | ermB | ColE1 | Clostridium ljungdahlii, Clostridium autoethanogenum | [54,59] |
| Species | Genetic Manipulations | Ref. |
|---|---|---|
| ClosTron | ||
| Clostridiumautoethanogenum | Disruption of [FeFe]-hydrogenase and [NiFe]-hydrogenase genes involved in energy conservation | [36] |
| Clostridiumautoethanogenum | Disruption of PCK, GAPDH, and Nfn complex genes | [37] |
| Clostridiumautoethanogenum | Disruption of acsA, cooS1, and cooS2 involved in carbon fixation | [38] |
| Clostridiumautoethanogenum | Disruption of adhE1, adhE2, aor1, and aor2 improved ethanol production to 180% | [28] |
| Clostridiumljungdahlii | Disruption of adhE1 reduced ethanol production | [39] |
| Transposon mutagenesis | ||
| Acetobacterium woodii | Insertion of Tn925 or Tn916 from E. faecalis | [63] |
| Clostridiumljungdahlii | Insertion of 5-kb acetone biosynthesis pathway via Himar1 transposase | [64] |
| Homologous recombination | ||
| Acetobacterium woodii | Deletion of ~5-kb rnfCDGEAB by HR in ∆pyrE mutant generated by allelic-coupled exchange | [25] |
| Acetobacterium woodii | Deletion of ~5-kb lctCDEF by HR in ∆pyrE mutant | [26] |
| Acetobacterium woodii | Deletion of ~3-kb hydBA by HR in ∆pyrE mutant | [27] |
| Clostridiumautoethanogenum | Double deletion of two aor or adhE isoforms via allelic exchange | [28] |
| Clostridiumljungdahlii | Deletion of rnfAB by HR | [29] |
| Clostridiumljungdahlii | Deletion of fliA via double-crossover; Deletion of adhE1 and adhE2 reduced ethanol production | [30] |
| Clostridiumljungdahlii | Insertion of a butyrate production pathway by HR; Deletion of pta, adhE1 and CLJU_c39430 via Cre-loxP system redirected carbon and electron flux from acetate to butyrate synthesis | [31] |
| Clostridiumljungdahlii | Insertion of a butyric acid production pathway by phage serine integrase | [32] |
| Moorellathermoacetica | Insertion of ldh by HR in ∆pyrF mutant | [33] |
| Thermoanaerobacter kivui | Deletion of fruK in ∆pyrE mutant | [34] |
| Thermoanaerobacter kivui | Deletion of fdhF, hycB3, hycB4, and hydA2 | [35] |
| CRISPR-Cas | ||
| Clostridiumautoethanogenum | SpCas9-mediated deletion of adh and 2,3-bdh | [40] |
| Clostridiumljungdahlii | SpCas9-mediated deletion of pta, adhE1, ctf and pyrE | [41] |
| Clostridiumljungdahlii | SpCas9-mediated insertion of attB site and elimination of specific sites | [32] |
| Clostridiumljungdahlii | FnCas12a-mediated deletion of pyrE, pta, adhE1, and ctf | [42] |
| Clostridiumljungdahlii | Single nucleotide substitution of pta, adhE1, adhE2, aor1, and aor2 using dCas9 fused with cytidine deaminase | [43] |
| Eubacterium limosum | SpCas9-mediated insertion of ermB gene into folD and acsC for gene disruption | [44] |
| Eubacterium limosum | SpCas9-mediated deletion of pyrF | [45] |
| Species | Plasmids | Genes | Product | Ref. |
|---|---|---|---|---|
| Clostridiumljungdahlii | pIMP1, pSOBPptb | thlA, bdhA, adhE | Butanol | [46] |
| Clostridiumljungdahlii | pMTL85246 | groES, groEL | Ethanol | [92] |
| Clostridiumautoethanogenum, Clostridiumljungdahlii | pMTL85245 | thlA, crt, bhd, bcd, etfAB | Butanol | [97] |
| Moorella thermoacetica | pBAD, pK18 | ldh, dpyrF, g3pd | Lactate | [33] |
| Clostridiumaceticum | pIMP1 | thlA, ctfA/B, adc, atoDA, teII, ybgC | Acetone | [47] |
| Clostridiumautoethanogenum, Clostridiumljungdahlii | pMTL85147 | thla, ctfA/B, adc | Acetone, Isopropanol | [96] |
| Clostridiumautoethanogenum | pMTL85245, pMTL83245 | malonyl-CoA reductase | 3-HP | [57] |
| Clostridiumautoethanogenum | pMTL85145, pMTL83155 | als, aldc, budC, adh, pddABC | 2,3-BDO, 2-Butanol | [93] |
| Clostridiumautoethanogenum | pMTL85146, pMTL85246 | ispS, idi, dxs, hmgs, mk, pmk, pmd | mevalonate, isoprene | [98] |
| Clostridiumautoethanogenum | pMTL85141, pMTL85241 | budA, adh, alsS | Acetoin, 2,3-BDO | [59] |
| Acetobacteriumwoodii | pJIR750ai | pta, ack, thf | Acetate | [50] |
| Clostridiumljungdahlii | pAH2, pKRAH1, pCL2, pJIR-ermB | adhE1, adhE2, thlA-ctfAB-adc | Ethanol, acetate, Acetone | [90] |
| Clostridiumljungdahlii | pMCSs, pJF100s, pDWs | ispS, idi | mevalonate, isoprene | [102] |
| Acetobacteriumwoodii | pMTL84151, pJIR750ai | thlA, ctfA/B, adc | Acetone | [51] |
| Clostridiumautoethanogenum | pMTL83157 | codh/acs | Ethanol, lactate | [86] |
| Clostridiumljungdahlii | pIMP1, pXY1 | thl, dnaK | Ethanol, acetate | [49] |
| Moorella thermoacetica | pK18 | aldh | Ethanol, acetate | [55] |
| Clostridiumljungdahlii | pMTL83151, pJF100s | ispS, idi | mevalonate, isoprene | [56] |
| Clostridiumautoethanogenum | pMTL83157 | phaA, phaB, phaC | PHB | [101] |
| Clostridiumljungdahlii, Clostridiumautoethanogenum | pMTLs, pANNE99 | panBCD, bioBDF, bioHCA | Pantothenate, Biotin, Thiamine | [103] |
| Acetobacteriumwoodii | pMTL83151 | gusA, arcABCD | Ornithine | [103] |
| Eubacterium limosum | pJIR750ai | gcvTH, gcvPA/B, grdX, trxAB, grdABCDE | Acetate | [52] |
| Eubacterium limosum | pJIR750ai | alsSD | Acetoin | [53] |
| Year | Specices | Omics-Study | Ref. |
|---|---|---|---|
| 2011. | Clostridiumljungdahlii, Clostridiumautoethanogenum, Clostridiumragsdalei | Genome | [7] |
| 2013 | Clostridiumljungdahlii | GEMs | [118] |
| 2013 | Clostridiumljungdahlii | Transcriptome | [105] |
| 2014 | Clostridiumautoethanogenum | Genome | [119] |
| 2015 | Clostridium aceticum | Genome | [120] |
| 2015 | Clostridiumautoethanogenum | Transcriptome | [36] |
| 2015 | Clostridiumautoethanogenum | Genome, GEMs, transcriptomics, metabolomics, Proteomics | [37] |
| 2015 | Clostridiumautoethanogenum | Transcriptome | [106] |
| 2015 | Clostridiumautoethanogenum | Genome | [121] |
| 2015 | Clostridiumljungdahlii | Transcriptome | [107] |
| 2016 | 14 species | Pan-Genome | [122] |
| 2016 | Clostridiumljungdahlii | GEMs | [89] |
| 2016 | Clostridiumljungdahlii, Clostridiumautoethanogenum, Clostridium ragsdalei, Clostridium coskatii | Comparison of genome | [39] |
| 2017 | Eubacterium limosum | Genome, TSS | [108] |
| 2017 | Clostridium autoethanogenum | GEMs, transcriptome | [113] |
| 2017 | Clostridium ljungdahlii | Transcriptome | [123] |
| 2018 | Acetobacteriumbakii | Genome, TSS, Transcriptome | [110] |
| 2018 | Eubacterium limosum | Transcriptome, Translatome | [112] |
| 2018 | Acetobacteriumwoodii | Transcriptome | [109] |
| 2018 | Clostridium ljungdahlii | Translatome | [111] |
| 2018 | Clostridium autoethanogenum | GEMs, Proteomics, Metabolomics | [124] |
| 2019 | Acetobacteriumpaludosum, Acetobacteriumtundrae, Acetobacteriumbakii, Alkalibaculum bacchi | Comparison of genome | [125] |
| 2019 | Clostridium autoethanogenum | GEMs | [114] |
| 2019 | Clostridium ljungdahlii | GEMs, Proteomics | [115] |
| 2019 | Clostridium sp. AWRP | Genome | [126] |
| 2020 | Clostridium drakei | Genome, Transcriptome, GEMs | [52] |
| 2020 | Clostridium autoethanogenum | GEMs, Proteomics, Metabolomics | [91] |
| 2020 | Clostridium ljungdahlii | GEMs, Transcriptome | [116] |
| 2020 | Clostridium autoethanogenum | GEMs | [117] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jin, S.; Bae, J.; Song, Y.; Pearcy, N.; Shin, J.; Kang, S.; Minton, N.P.; Soucaille, P.; Cho, B.-K. Synthetic Biology on Acetogenic Bacteria for Highly Efficient Conversion of C1 Gases to Biochemicals. Int. J. Mol. Sci. 2020, 21, 7639. https://doi.org/10.3390/ijms21207639
Jin S, Bae J, Song Y, Pearcy N, Shin J, Kang S, Minton NP, Soucaille P, Cho B-K. Synthetic Biology on Acetogenic Bacteria for Highly Efficient Conversion of C1 Gases to Biochemicals. International Journal of Molecular Sciences. 2020; 21(20):7639. https://doi.org/10.3390/ijms21207639
Chicago/Turabian StyleJin, Sangrak, Jiyun Bae, Yoseb Song, Nicole Pearcy, Jongoh Shin, Seulgi Kang, Nigel P. Minton, Philippe Soucaille, and Byung-Kwan Cho. 2020. "Synthetic Biology on Acetogenic Bacteria for Highly Efficient Conversion of C1 Gases to Biochemicals" International Journal of Molecular Sciences 21, no. 20: 7639. https://doi.org/10.3390/ijms21207639